Maciej Mazur
on 10 December 2021
Windows Subsystem for Linux for data scientists
Ubuntu is the number one choice for data scientists worldwide. It is also by far the most popular Linux distribution used on public clouds with machine learning offerings. However, we don’t forget about our Windows friends – many of whom had their operating system chosen by corporate IT departments. Windows users can still get all the benefits of using Ubuntu thanks to Windows Subsystem for Linux.
This post will walk you through a complete setup from the new Windows 10 installation to a complete data scientist work environment. The assumption is that you have a laptop / desktop with Windows 10 and NVidia GPU. What we want to achieve is a stack like this:
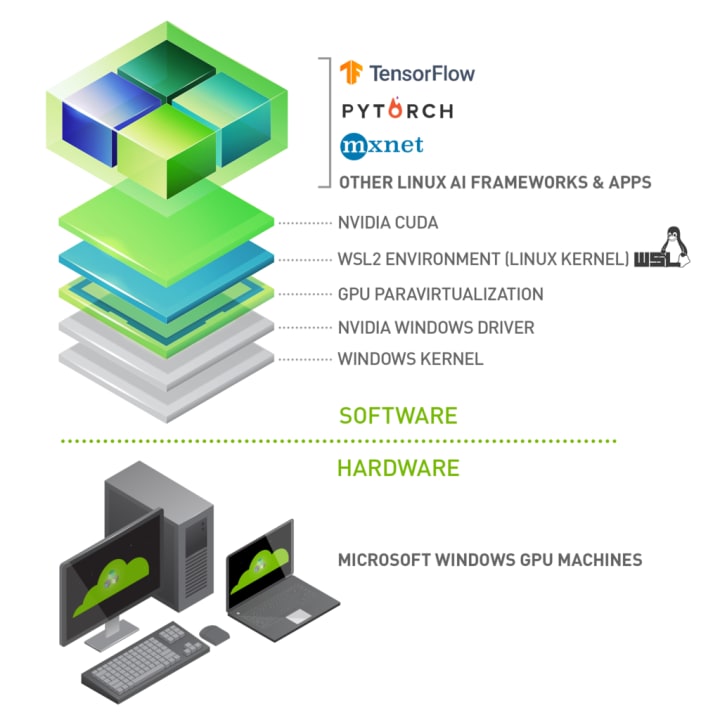
Step 1: Install WSL
First, we need to start by installing WSL itself.
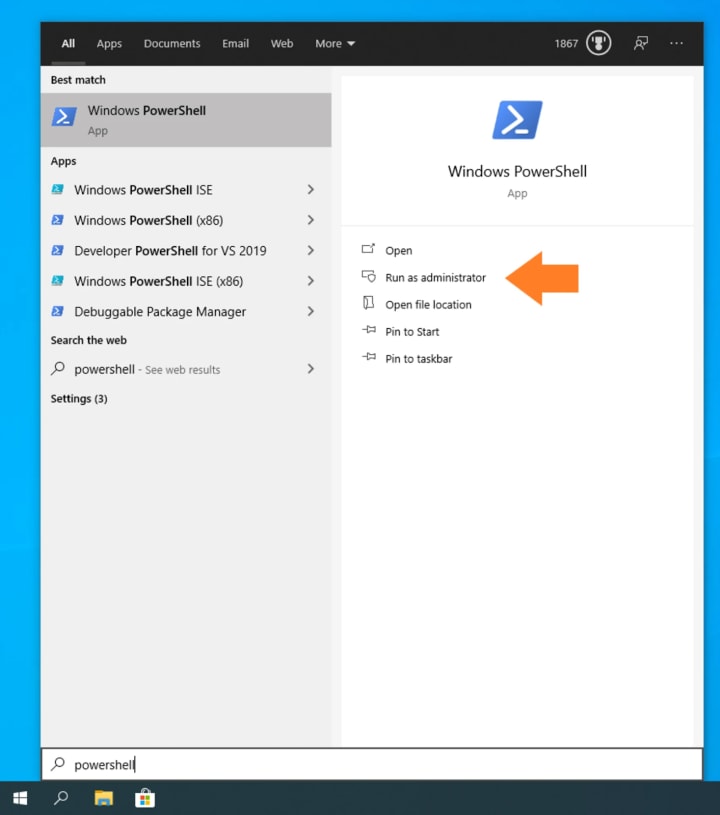
Open PowerShell as an Administrator:
Type the following command to enable WSL:
dism.exe /online /enable-feature /featurename:VirtualMachinePlatform /all /norestart
Restart your computer.
After restarting, download and install the WSL 2 Linux kernel from Microsoft for your device architecture:
Finally, we recommend you to set WSL 2 as the default WSL environment:
wsl.exe –set-default-version 2
Step 2: Install Ubuntu
Download Ubuntu for WSL from the Microsoft Store.
Run Ubuntu from the Start menu.
Select a username and password for your administrative user.
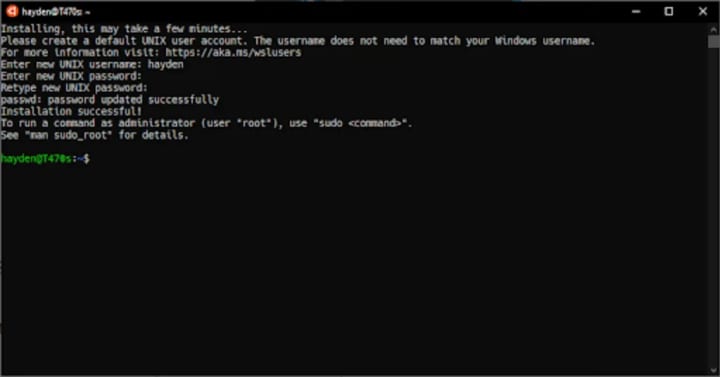
Once the Ubuntu installation is complete, we recommend you download and try the new Windows Terminal for the best Ubuntu on WSL experience.
Step 3: Install GPU drivers and Docker
Next, install the proper GPU drivers. Depending on your GPU, choose the right package from https://developer.nvidia.com/cuda/wsl/download. There is no need to install the driver inside WSL VMs, as they use the driver installed in Windows.
After that, open the Windows terminal, and set up Docker inside your Ubuntu installation:
curl https://get.docker.com | sh
distribution=$(. /etc/os-release;echo $ID$VERSION_ID)
curl -s -L https://nvidia.github.io/nvidia-docker/gpgkey | sudo apt-key add –
curl -s -L https://nvidia.github.io/nvidia-docker/$distribution/nvidia-docker.list | sudo tee /etc/apt/sources.list.d/nvidia-docker.list
sudo apt-get update
sudo apt-get install -y nvidia-docker2
sudo service docker stop
sudo service docker start
Now, check that everything is working fine by running a benchmark:
docker run –gpus all nvcr.io/nvidia/k8s/cuda-sample:nbody nbody -gpu -benchmark
Step 4: Install Data Science Stack
After this, install the Data Science Stack:
git clone https://github.com/NVIDIA/data-science-stack
cd data-science-stack
./data-science-stack setup-system
This allows you to now create and run containers with all necessary tools:
./data-science-stack list
./data-science-stack build-container
./data-science-stack run-container
The reverse of build-container is purge-container.
Step 5: Enjoy!
With everything set up, youcan now go to http://localhost:8888/ and enjoy the Jupyter notebook and full power of Linux CLI.
You can try this, for example, with HP Z series laptops, which come with a pre-setup environment so that you can start working just after the first boot of your new machine.
What’s next
There are many more reasons why Ubuntu is the de-facto standard operating system for data science. You can find them all on https://ubuntu.com/ai



